Multiprocessing¶
Processing magnetotelluric field surveys can be demanding if there many sites and long recordings. To help speed up processing, resistics supports multiprocessor computation using the python multiprocessing module (read more here). Multiprocessing can be used in the following processing steps:
Spectra calculation
Statistic calculation
Transfer function estimation (robust regression)
Important
For Windows users, multiprocessing requires scripts using resistics and multiprocessing to be protected using if __name__ == ‘__main__’. For a more detailed explanation of why, please see the offical python documentation. What this means is illustrated in the below example.
Multiprocessing is activated by specifying the ncores keyword in the following methods:
processProject()orprocessSite()
The optional argument ncores is expected to be an integer. It is recommend to use less cores than cores available on the system. This will leave spare processing power for system tasks and for interaction with the system.
Alternatively and probably more usefully, the ncores options can be set in configuration files, where it can be either a subparameter or a global parameter.
For example, setting ncores as a global parameter:
1 2 3 4 5 6 7 8 9 | name = multi
ncores = 6
[Spectra]
specdir = "multi"
[Statistics]
stats = "coherence", "transferFunction",
remotestats = "RR_coherenceEqn", "RR_transferFunction"
|
By setting a global ncores option, any methods that support multiprocessing will use it on the globally specified number of cores.
Another option is to define ncores separately for individual tasks. The ncores option can be supplied to the following sections separately.
Spectra
Statistics
Solver
The following configuration specifies different cores for each section.
1 2 3 4 5 6 7 8 9 10 | name = multi
[Spectra]
ncores = 4
specdir = "multi2"
[Statistics]
ncores = 5
stats = "coherence", "transferFunction",
remotestats = "RR_coherenceEqn", "RR_transferFunction"
|
In the above config file, spectra calculations will use 4 cores, statistic calculations will use 5 cores and the solver (robust regression) will use only a single core.
Example¶
The below configuration and processing script runs spectra, statistic and transfer function calculations on 6 cores.
1 2 3 4 5 6 7 8 9 | name = multi
ncores = 6
[Spectra]
specdir = "multi"
[Statistics]
stats = "coherence", "transferFunction",
remotestats = "RR_coherenceEqn", "RR_transferFunction"
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
from resistics.project.spectra import calculateSpectra
from resistics.project.transfunc import processProject, viewImpedance
from resistics.project.statistics import calculateStatistics
from resistics.project.mask import newMaskData, calculateMask
if __name__ == "__main__":
projData = loadProject(projectPath, "multiconfig.ini")
# calculate spectrum using standard options
calculateSpectra(projData)
projData.refresh()
calculateStatistics(projData)
# process project with standard options
processProject(projData)
figs = viewImpedance(projData, oneplot=False, show=False, save=False)
figs[0].savefig(imagePath / "multproc_standard_process")
# calculate mask for 128
maskData = newMaskData(projData, 128)
maskData.setStats(["coherence"])
maskData.addConstraint("coherence", {"cohExHy": [0.7, 1.0], "cohEyHx": [0.7, 1.0]})
maskData.maskName = "coh70_100"
calculateMask(projData, maskData, sites=["site1"])
# calculate mask for 4096
maskData = newMaskData(projData, 4096)
maskData.setStats(["coherence"])
maskData.addConstraint("coherence", {"cohExHy": [0.7, 1.0], "cohEyHx": [0.7, 1.0]})
maskData.maskName = "coh70_100"
calculateMask(projData, maskData, sites=["site1"])
# process with masks
processProject(projData, masks={"site1": "coh70_100"}, postpend="coh70_100")
figs = viewImpedance(
projData, oneplot=False, postpend="coh70_100", show=False, save=False
)
figs[0].savefig(imagePath / "multproc_mask_process")
|
Warning
This script was run on the Windows platform. Note the use of if __name__ == ‘__main__’ to ensure no multiprocessing issues are encountered. Forgetting this part would cause many processes to be spawned which will consume system resources.
The impedance tensor estimates are shown below with and without masking.
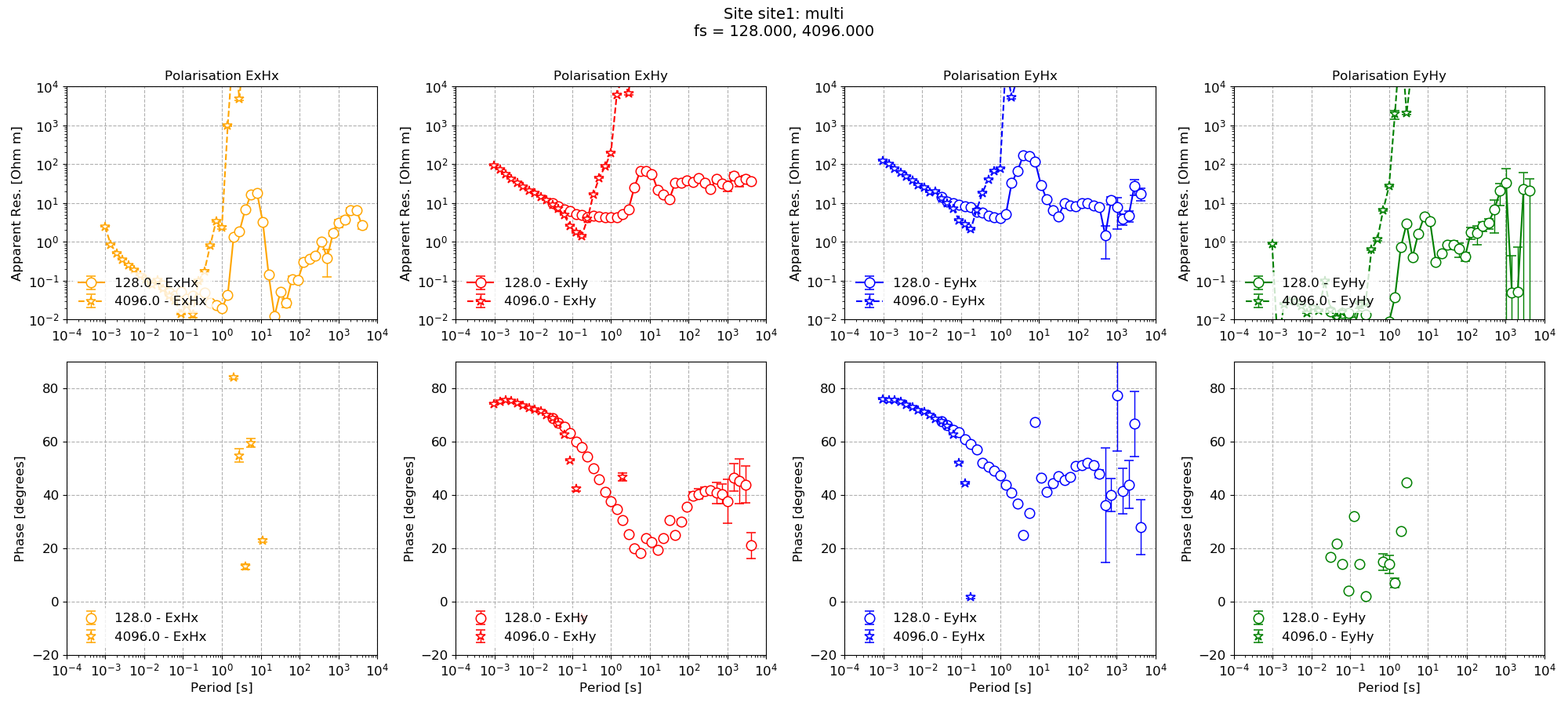
Multiprocessing using all standard options¶
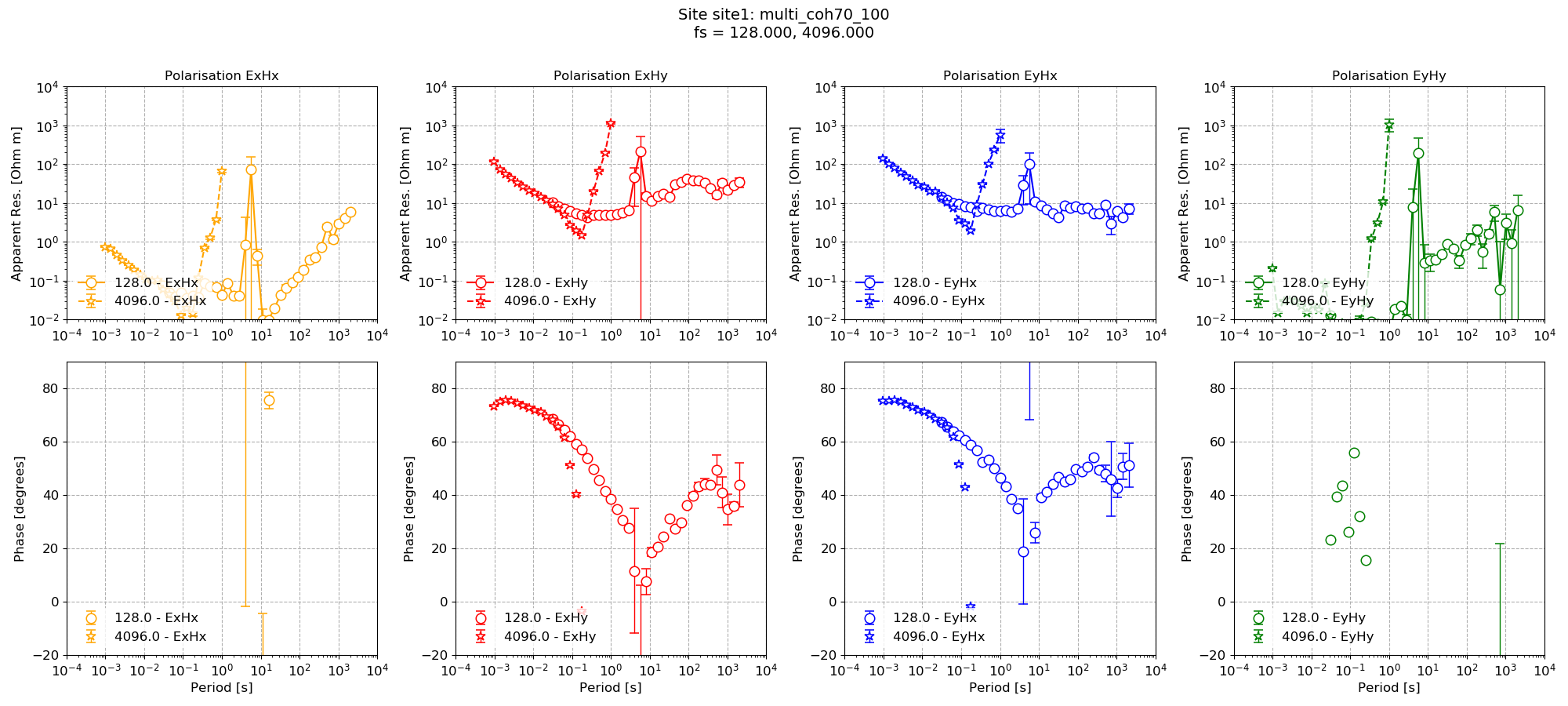
Multiprocessing with masks¶
Complete example script¶
For the purposes of clarity, the complete example script is provided below.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
from resistics.project.spectra import calculateSpectra
from resistics.project.transfunc import processProject, viewImpedance
from resistics.project.statistics import calculateStatistics
from resistics.project.mask import newMaskData, calculateMask
if __name__ == "__main__":
projData = loadProject(projectPath, "multiconfig.ini")
# calculate spectrum using standard options
calculateSpectra(projData)
projData.refresh()
calculateStatistics(projData)
# process project with standard options
processProject(projData)
figs = viewImpedance(projData, oneplot=False, show=False, save=False)
figs[0].savefig(imagePath / "multproc_standard_process")
# calculate mask for 128
maskData = newMaskData(projData, 128)
maskData.setStats(["coherence"])
maskData.addConstraint("coherence", {"cohExHy": [0.7, 1.0], "cohEyHx": [0.7, 1.0]})
maskData.maskName = "coh70_100"
calculateMask(projData, maskData, sites=["site1"])
# calculate mask for 4096
maskData = newMaskData(projData, 4096)
maskData.setStats(["coherence"])
maskData.addConstraint("coherence", {"cohExHy": [0.7, 1.0], "cohEyHx": [0.7, 1.0]})
maskData.maskName = "coh70_100"
calculateMask(projData, maskData, sites=["site1"])
# process with masks
processProject(projData, masks={"site1": "coh70_100"}, postpend="coh70_100")
figs = viewImpedance(
projData, oneplot=False, postpend="coh70_100", show=False, save=False
)
figs[0].savefig(imagePath / "multproc_mask_process")
|