Using configuration files¶
Configuration files are a powerful way to set multiple parameters. For more information about the parameters in configuration files, please see Configuration files.
A user supplied configuration only needs to include non-default parameters (those where the values are changed). Details on the easiest way to create a configuration file are provided here.
To use configuration files, they should be specified when loading in a project. An example configuration file is shown below.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | name = dec8_5
[Spectra]
specdir = "dec8_5"
[Decimation]
numlevels = 8
[Frequencies]
perlevel = 5
[Window]
minwindowsize = 256
minoverlapsize = 64
|
This configuration has been named dec8_5 because it uses a decimation scheme of 8 decimation levels with 5 evaluation frequencies per level. Configuration names should be meaningful and unique to allow disambiguation and easy repeatability. Configuration file names are stored in dataset comments to ensure transparency.
The below shows how to load a project with a configuration specified by a configuration file.
1 2 3 4 5 6 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load the project and also provide a config file
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
projData.printInfo()
|
The configuration file above specifies a spectra directory to use. For more on what this means, please see the Multiple spectra page. The result is that all project methods which use the ProjectData instance loaded with the configuration file will pick up the configuration parameters.
Calculating spectra is now straightforward. When passing the ProjectData instance loaded with the tutorialConfig.ini configuration file to calculateSpectra(), the specdir option in the configuration file will automatically be picked up.
8 9 10 11 12 | # calculate spectrum using the new configuration
from resistics.project.spectra import calculateSpectra
calculateSpectra(projData)
projData.refresh()
|
The spectra will be stored in the following location:
exampleProject
├── calData
├── timeData
│ └── site1
| |── dataFolder1
│ |── dataFolder2
| |── .
| |── .
| |── .
| └── dataFolderN
├── specData
│ └── site1
| |── dataFolder1
| | └── dec8_5
| |
│ |── dataFolder2
| | └── dec8_5
| |── .
| |── .
| |── .
| └── dataFolderN
| └── dec8_5
├── statData
├── maskData
├── transFuncData
├── images
└── mtProj.prj
To process these spectra, it’s a similar situation. Passing the ProjectData instance loaded with the tutorialConfig.ini configuration file to processProject(), the specdir option in the configuration file will automatically be picked up.
14 15 16 17 18 19 | # process the spectra
from resistics.project.transfunc import processProject, viewImpedance
processProject(projData)
figs = viewImpedance(projData, sites=["site1"], oneplot=False, save=False, show=False)
figs[0].savefig(imagePath / "usingConfigFiles_viewimp")
|
Now the plot using the new configuration parameters (8 decimation levels with 5 evaluation frequencies per decimation level) can be compared to the default parameterisation.
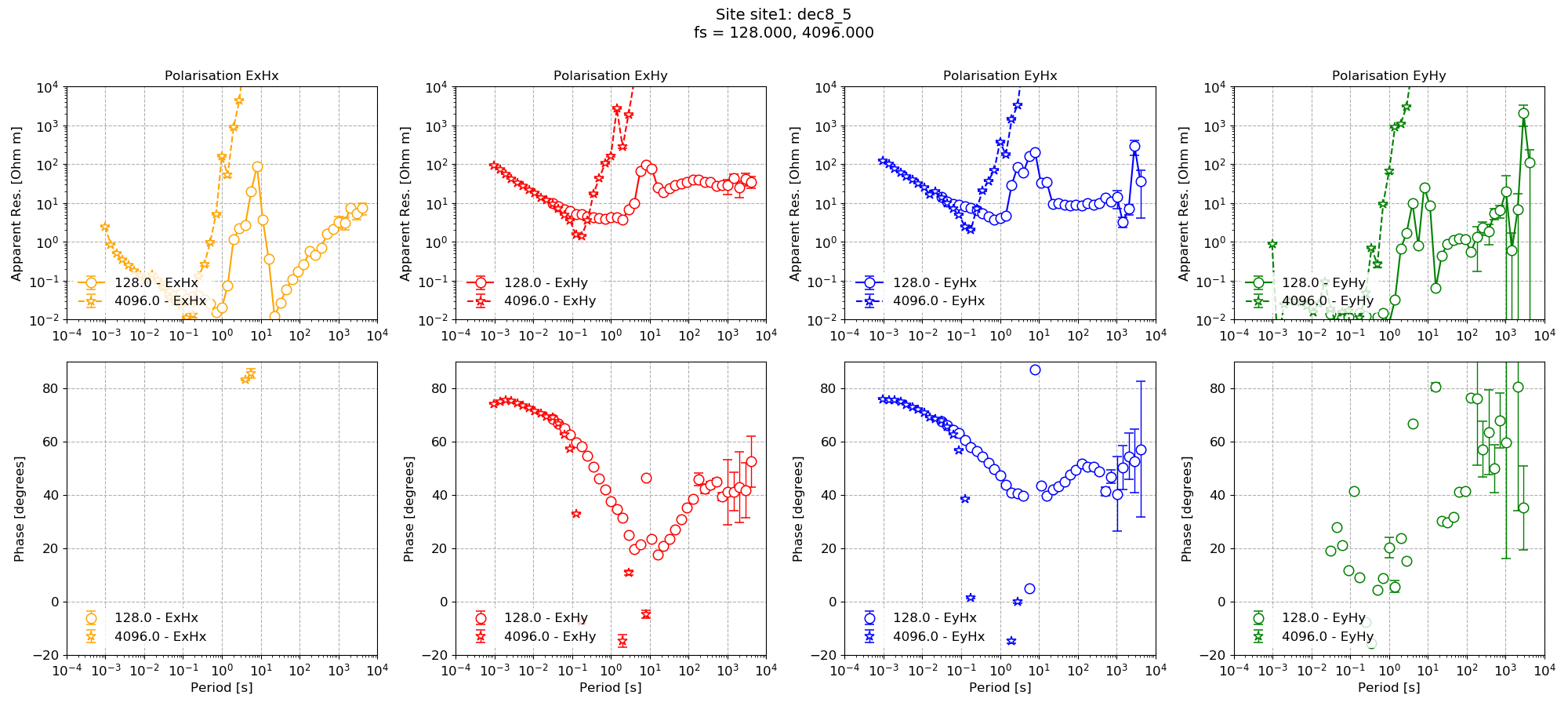
Using the customised configuration¶
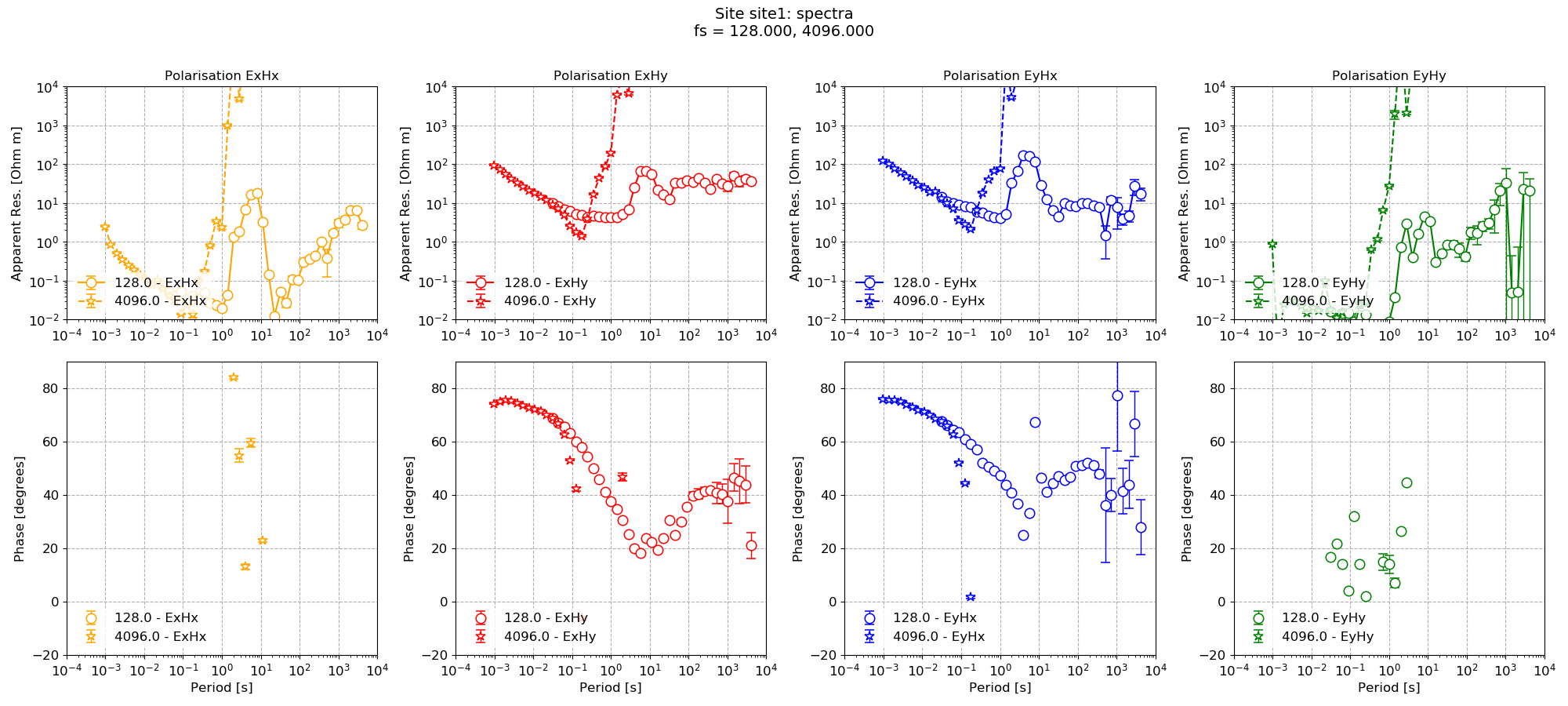
Using the default configuration¶
Complete example script¶
For the purposes of clarity, the complete example script is provided below.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load the project and also provide a config file
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
projData.printInfo()
# calculate spectrum using the new configuration
from resistics.project.spectra import calculateSpectra
calculateSpectra(projData)
projData.refresh()
# process the spectra
from resistics.project.transfunc import processProject, viewImpedance
processProject(projData)
figs = viewImpedance(projData, sites=["site1"], oneplot=False, save=False, show=False)
figs[0].savefig(imagePath / "usingConfigFiles_viewimp")
|