Processing with masks¶
Now that statistics and masks have been calculated, the final step is to include those masks in the transfer function calculation. There is a simple addition to the standard transfer function calculation workflow presented in Up and running and Tippers.
As always, load the project.
1 2 3 4 5 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load project and configuration file
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
|
Now perform the transfer function calculation with the addition of a mask.
7 8 9 10 11 12 13 14 15 16 17 | # Process the data with this mask
from resistics.project.transfunc import processProject
processProject(
projData,
sampleFreqs=[4096, 128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100"},
postpend="coh70_100",
)
|
The elements of note in this code block are:
masks keyword. A mask needs to be associated with a site and provided in a dictionary. More than a single mask file can be supplied for a site by passing a list of masks.
postpend keyword. This is the postpend on the output transfer function data file to avoid overwriting existing transfer function calculation files.
The resultant impedance tensor result can be visualised in the same way as demonstrated in previous sections using the viewImpedance() method of the project transfunc module.
1 2 3 4 5 6 7 8 9 10 11 | from resistics.project.transfunc import viewImpedance
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "runWithMask_coh70_100")
|
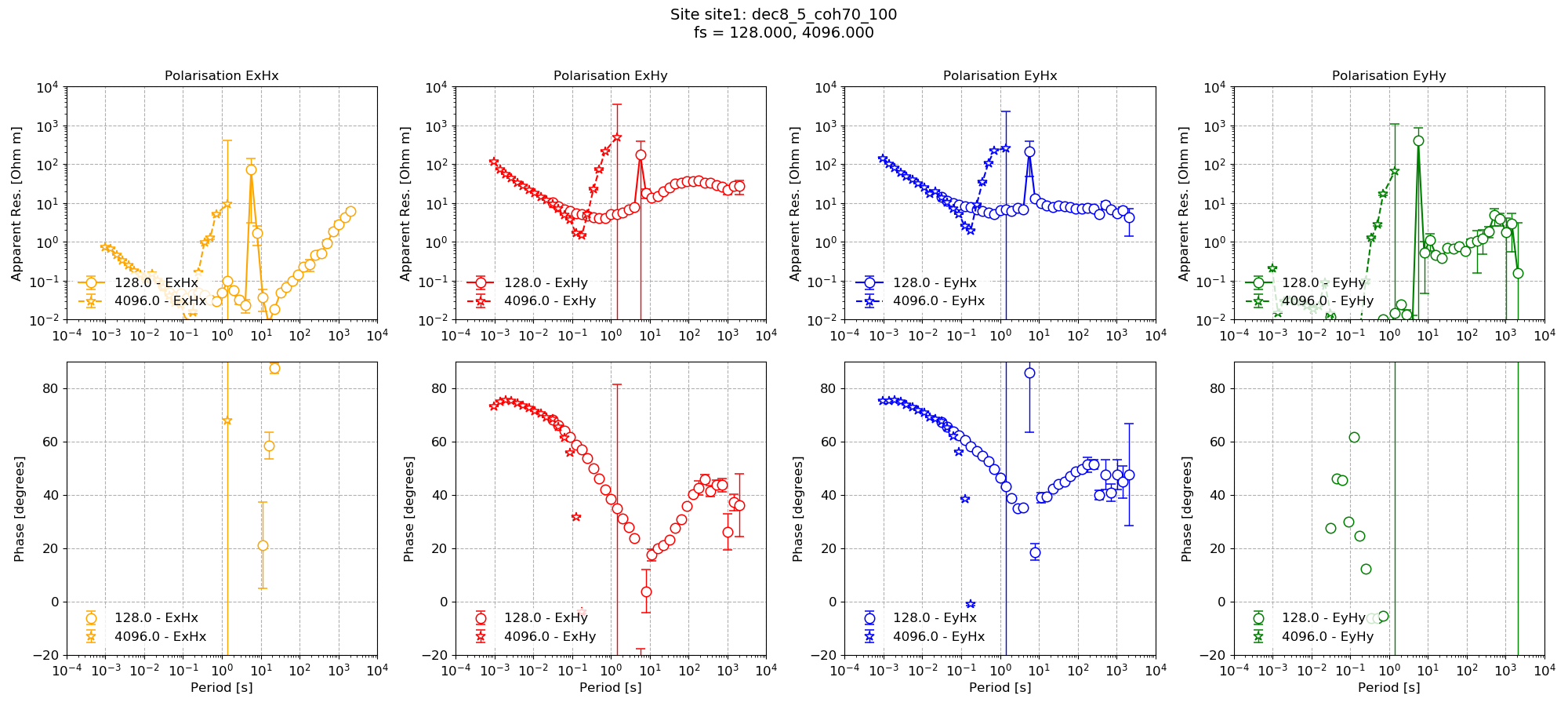
Impedance tensor estimate with the coherence masking¶
Repeating the process for the mask with additional transfer function constraints gives:
31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 | processProject(
projData,
sampleFreqs=[4096, 128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100_tfConstrained"},
postpend="coh70_100_tfConstrained",
)
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100_tfConstrained",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "runWithMask_coh70_100_tfConstrained")
|
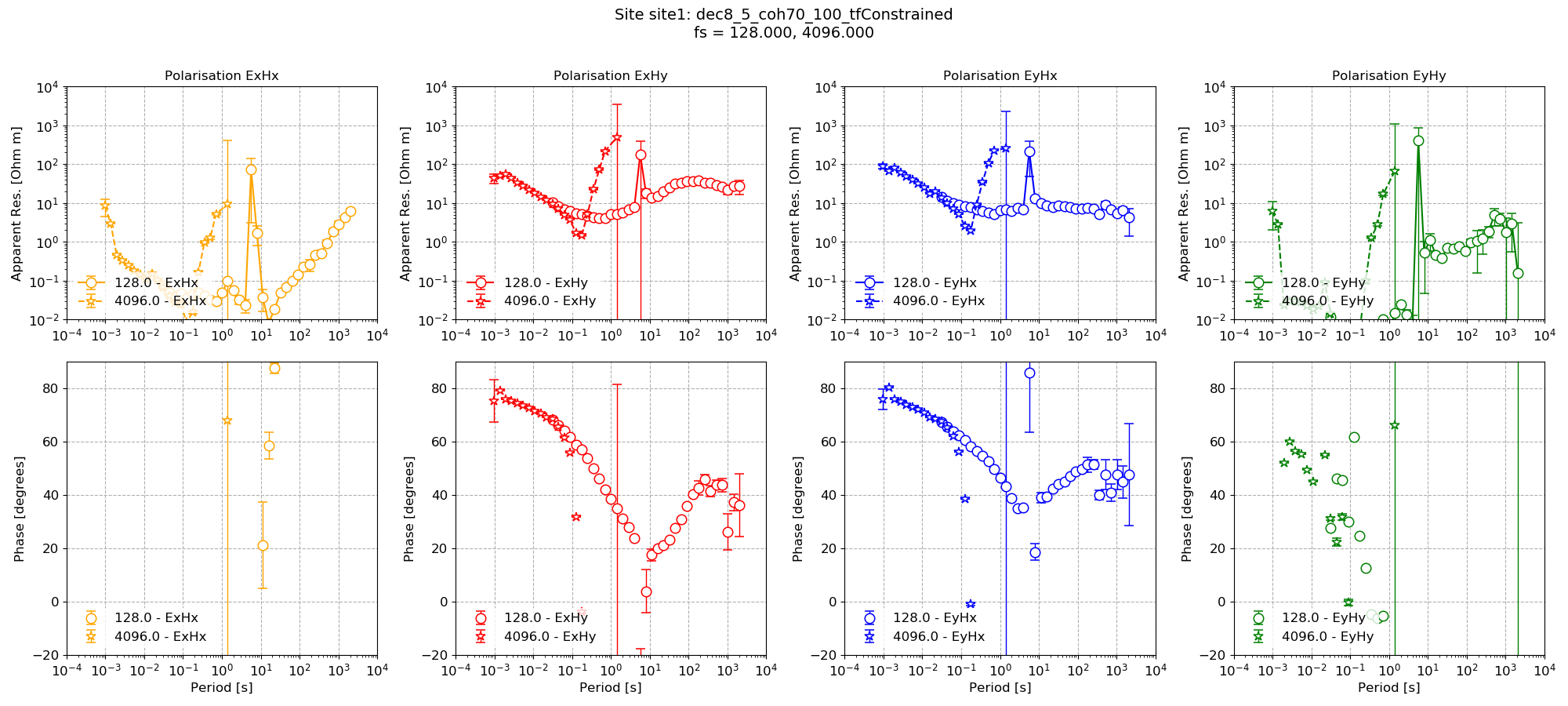
Impedance tensor estimate with both coherence and transfer function masking¶
Complete example script¶
For the purposes of clarity, the complete example script is provided below.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load project and configuration file
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
# Process the data with this mask
from resistics.project.transfunc import processProject
processProject(
projData,
sampleFreqs=[4096, 128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100"},
postpend="coh70_100",
)
from resistics.project.transfunc import viewImpedance
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "runWithMask_coh70_100")
processProject(
projData,
sampleFreqs=[4096, 128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100_tfConstrained"},
postpend="coh70_100_tfConstrained",
)
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100_tfConstrained",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "runWithMask_coh70_100_tfConstrained")
|