Date and time constraints¶
Another way of constraining the data before evaluation frequency calculation is through date and time constraints. In many cases, it can be advantageous to explicitly define the dates and times of the time windows (and their respective spectra) to include in the transfer function calculation.
There are three types of date and time constraints:
A time constraint which recurs on a daily basis
A date constraint to define dates to use
A combined date and time constraint
Note
For a better understanding of how window selection works, see the Recap and deeper dive.
As always, the means of providing date and time constraints will be demonstrated through an example. Begin as always by loading the project.
1 2 3 4 5 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load the project
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
|
The next step is define a list to hold the date/time constraints and add a single date/time constraint, which is in the form of a dictionary.
7 8 9 10 11 | # define date/time constraints - only time windows within the constraints will be used
datetimes = list()
datetimes.append(
{"type": "datetime", "start": "2012-02-10 19:00:00", "stop": "2012-02-11 07:00:00"}
)
|
As stated above, there are three types of datetime constraints. They are each specified in the following way:
Time constraint as a dictionary: {“type”: “time”, “start”: “HH:MM:SS”, “stop”: “HH:MM:SS”}. If the stop time is less than the start time, the time constraint is assumed to cross days. For example, {“type”: “time”, “start”: “20:00:00”, “stop”: “07:00:00”} will take only night time data.
Date constraint as a dictionary: {“type”: “date”, “date”: “YYYY-MM-DD”}.
Datetime constraint as a dictionary: {“type”: “datetime”, “start”: “YYYY-MM-DD HH:MM:SS”, “stop”: “YYYY-MM-DD HH:MM:SS”}
Multiple date/time constraints can be provided and they will be combined and only windows which fall within these date/time constraints will be used. Below is an example of processing with date/time constraints.
13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 | # process the spectra
from resistics.project.transfunc import processProject
processProject(
projData, sampleFreqs=[128], datetimes=datetimes, postpend="datetimeConstraint"
)
# plot transfer function and save the plot
from resistics.project.transfunc import viewImpedance
figs = viewImpedance(
projData,
sites=["site1"],
postpend="datetimeConstraint",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "datetimeConstraints_viewimp")
|
There is little new in this example apart from the addition of the datetimes keyword to specify the date/time constraints.
The processing gives the resultant transfer function (note, only processing 128 Hz time series data here).
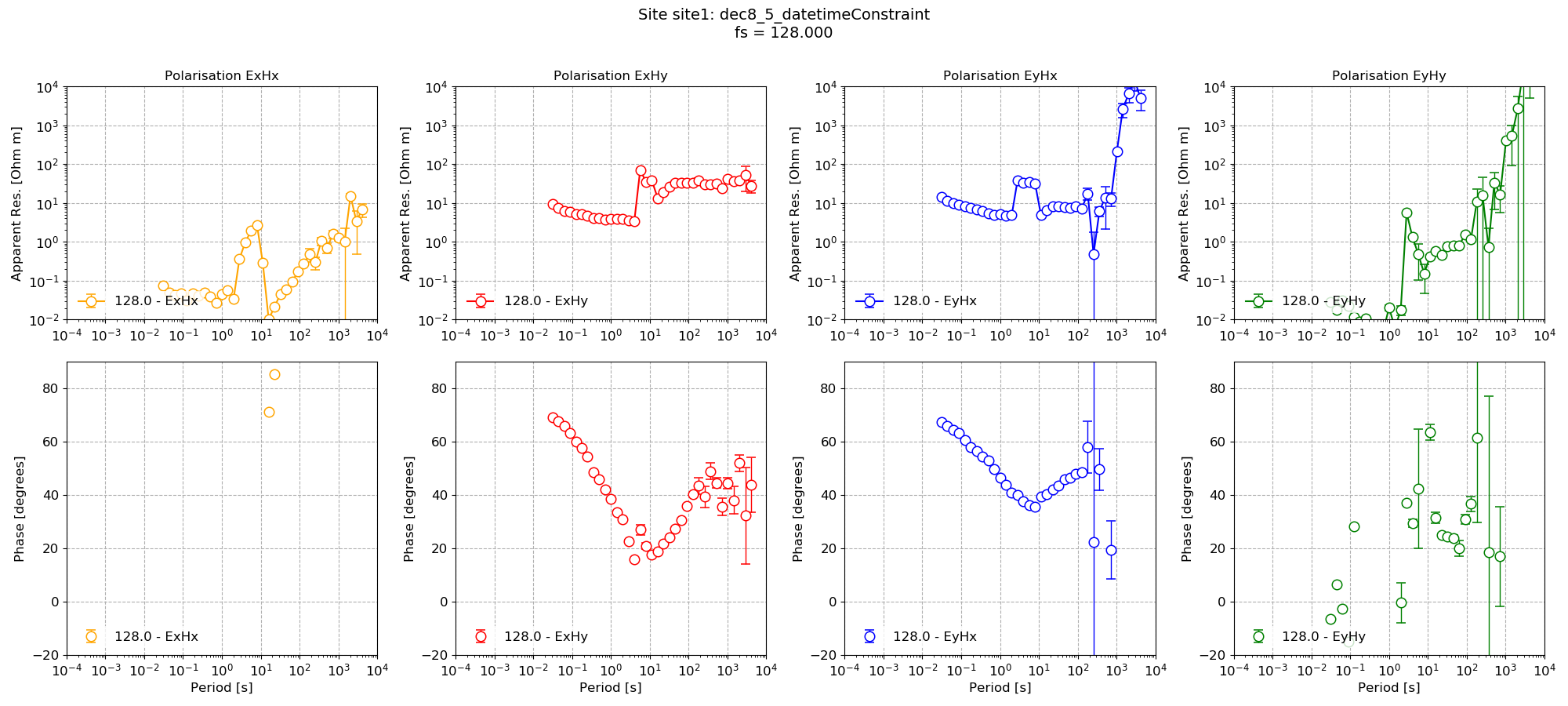
Impedance tensor estimate with date/time constraints¶
Compare this to the same processing but without datetime constraints.
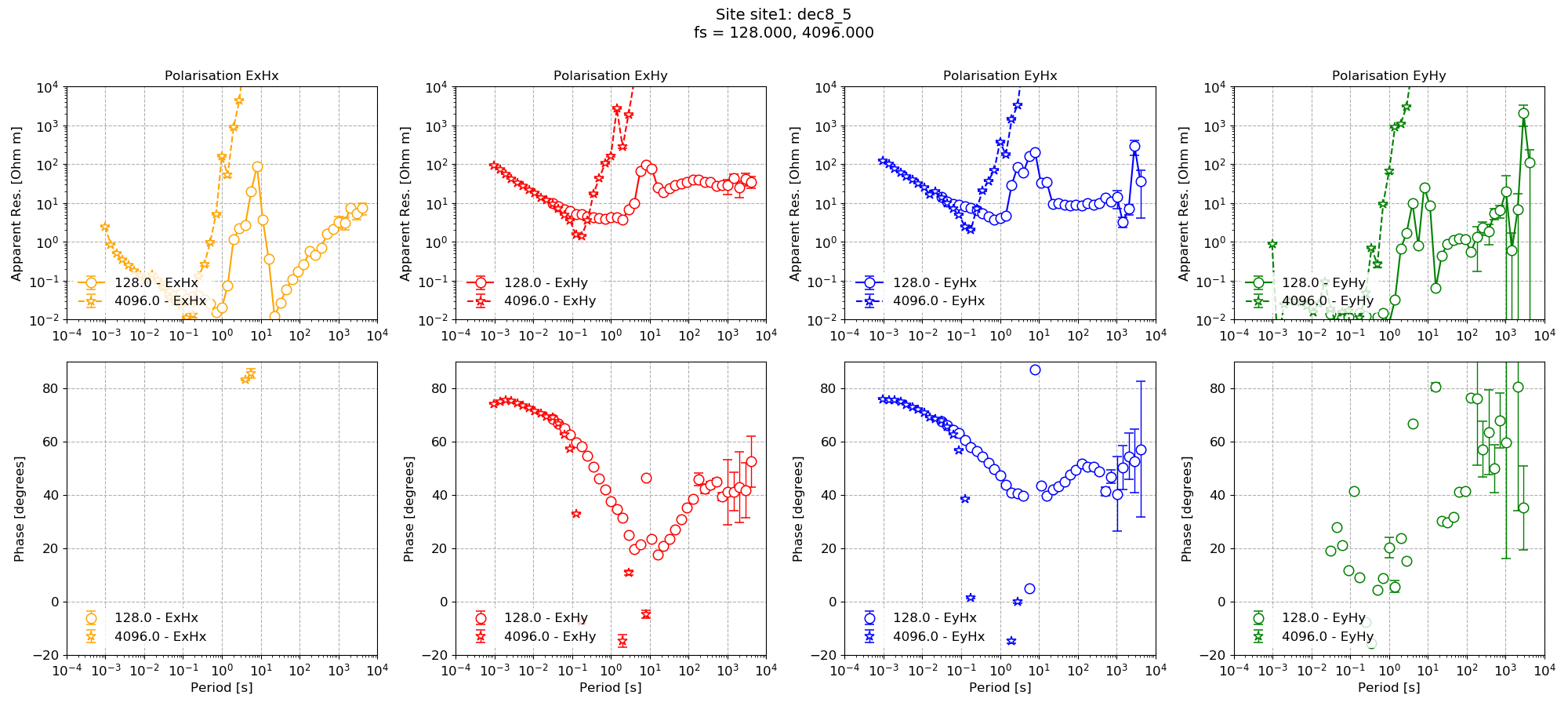
Impedance tensor estimate without date/time constraints¶
Finally, date/time constraints and masks can be combined as shown in the following example.
33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 | # process again with a mask too
processProject(
projData,
sampleFreqs=[128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100_tfConstrained"},
datetimes=datetimes,
postpend="coh70_100_tfConstrained_datetimeConstrained",
)
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100_tfConstrained_datetimeConstrained",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "datetimeConstraints_viewimp_mask_coh70_100_tfConstrained")
|
This gives the below transfer function.
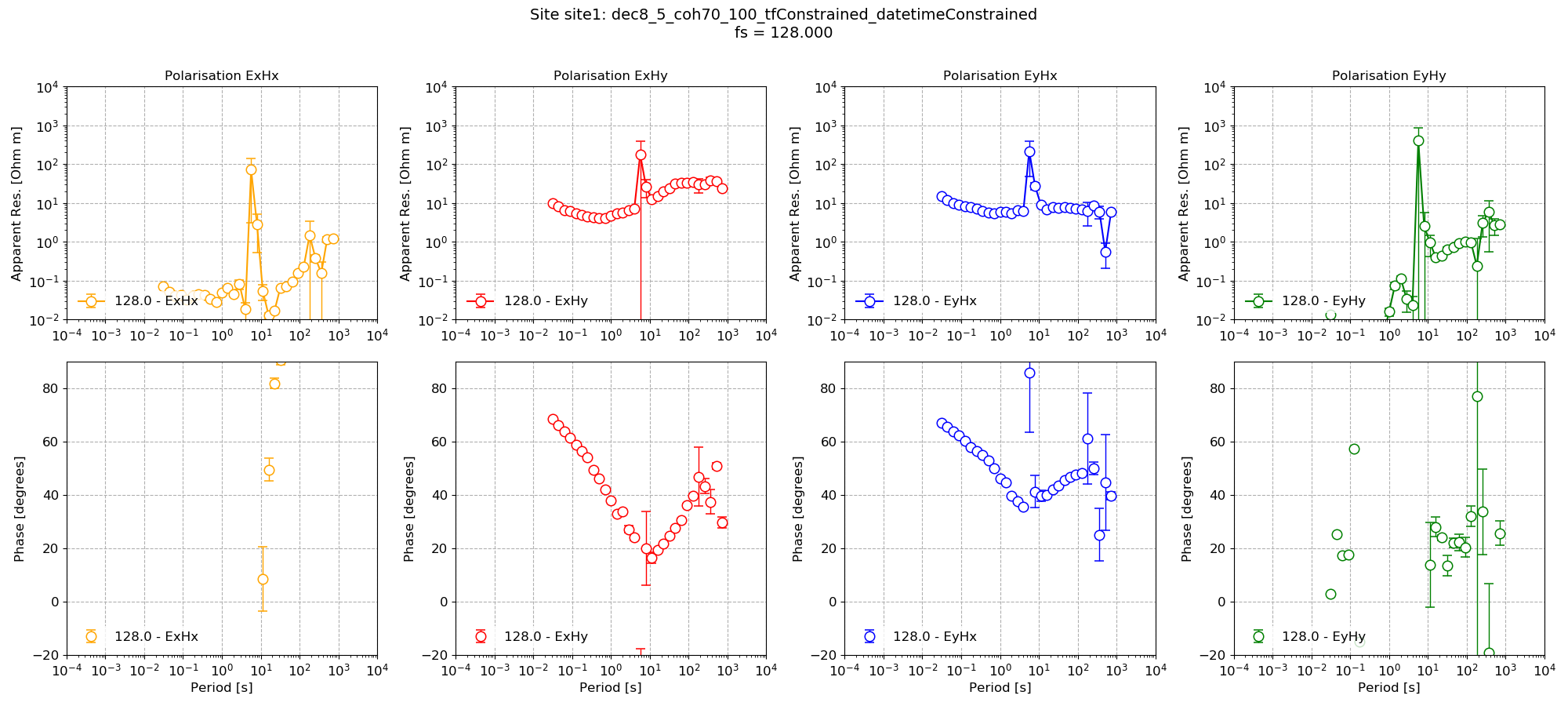
Impedance tensor estimate with date/time constraints and masks based on coherence and transfer function statistics¶
In comparison, the transfer function with just the mask.
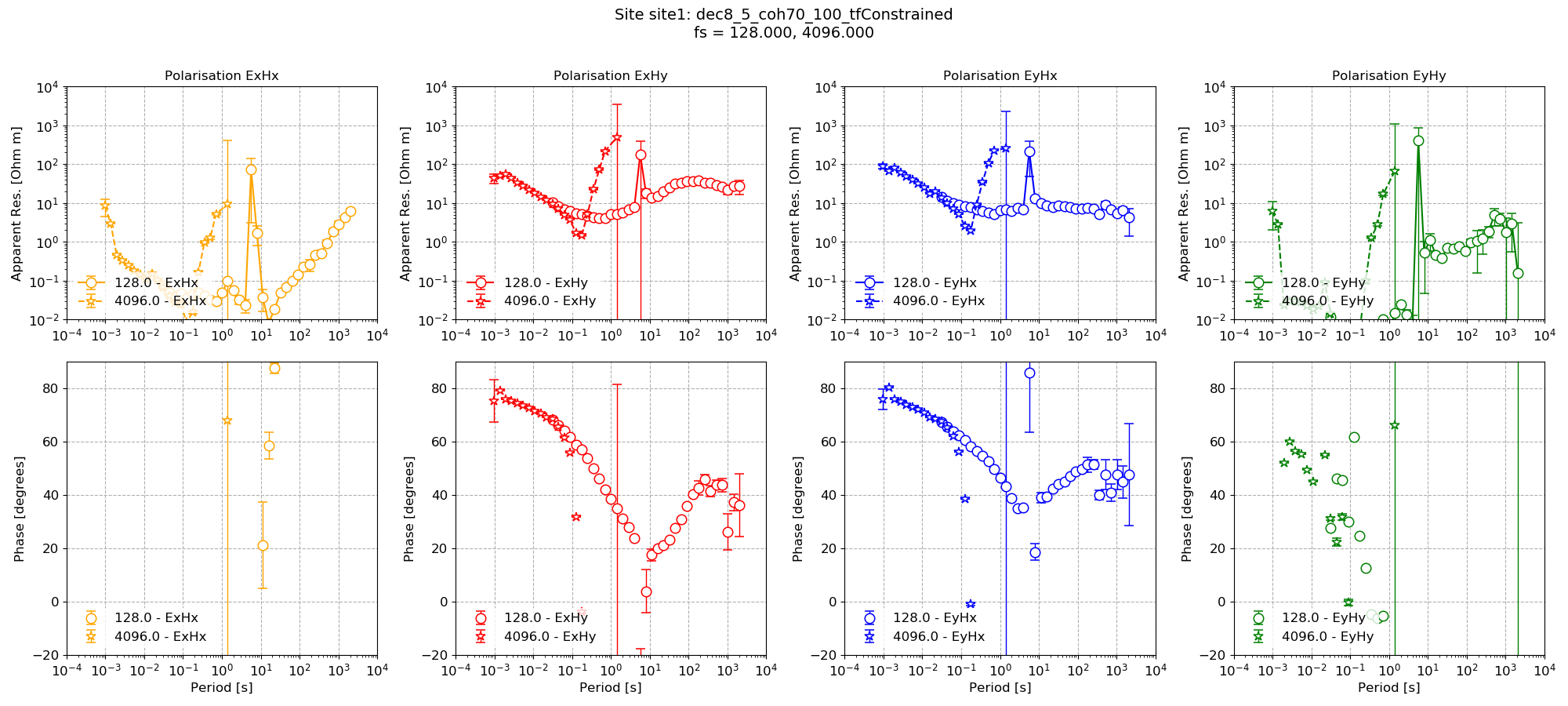
Impedance tensor estimate with just the masking based on coherence and transfer function statistics¶
Complete example script¶
For the purposes of clarity, the complete example script is provided below.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 | from datapaths import projectPath, imagePath
from resistics.project.io import loadProject
# load the project
projData = loadProject(projectPath, configFile="tutorialConfig.ini")
# define date/time constraints - only time windows within the constraints will be used
datetimes = list()
datetimes.append(
{"type": "datetime", "start": "2012-02-10 19:00:00", "stop": "2012-02-11 07:00:00"}
)
# process the spectra
from resistics.project.transfunc import processProject
processProject(
projData, sampleFreqs=[128], datetimes=datetimes, postpend="datetimeConstraint"
)
# plot transfer function and save the plot
from resistics.project.transfunc import viewImpedance
figs = viewImpedance(
projData,
sites=["site1"],
postpend="datetimeConstraint",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "datetimeConstraints_viewimp")
# process again with a mask too
processProject(
projData,
sampleFreqs=[128],
sites=["site1"],
outchans=["Ex", "Ey"],
masks={"site1": "coh70_100_tfConstrained"},
datetimes=datetimes,
postpend="coh70_100_tfConstrained_datetimeConstrained",
)
figs = viewImpedance(
projData,
sites=["site1"],
postpend="coh70_100_tfConstrained_datetimeConstrained",
oneplot=False,
save=False,
show=False,
)
figs[0].savefig(imagePath / "datetimeConstraints_viewimp_mask_coh70_100_tfConstrained")
|